Further Examples¶
In this section we present multiple examples for the mass conservation, semi-diffusive, and general approaches. In addition to this, we provide some examples satisfying the deficiency theorems. Before each example we depict the CellDesigner layout and C-graph generated using the instructions in Generating Presentable C-graphs. Those nodes that represent zero complexes are colored red while regular nodes are green.
Contents
- Further Examples
- Low Deficiency Approach
- Mass Conservation Approach
- Closed graph of Figure 5A of [OMYS17]
- Gene regulatory network with two mutually repressing genes from [OMYS14]
- Enzymatic reaction with inhibition by substrate from [OMBA09]
- Enzymatic reaction with simple substrate cycle from [HC87]
- G1/S transition in the cell cycle of Saccharomyces cerevisiae from [CFRS07]
- Double phosphorylation in signal transduction of [OGM+06]
- Double insulin binding
- p85-p110-PTEN
- Closed version of Figure 4B from [OMYS17]
- Closed version of Figure 4C from [OMYS17]
- Semi-diffusive Approach
- General Approach
Low Deficiency Approach¶
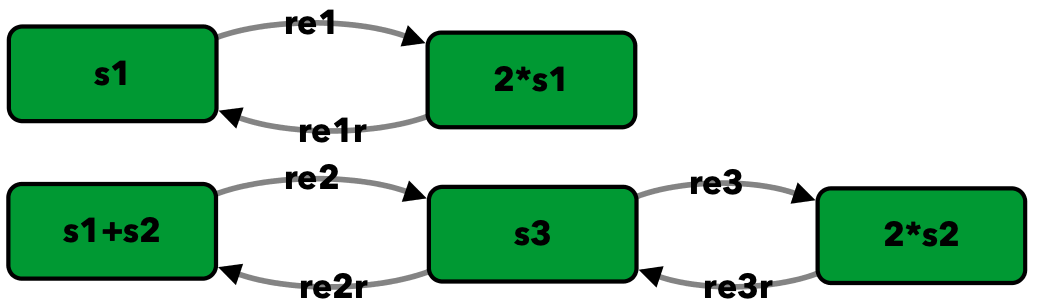
Network 3.13 of [Fei79]¶

To run this example download the SBML file and script
run_feinberg_ex3_13. After running this script we obtain
the following output:
Number of species: 3
Number of complexes: 5
Number of reactions: 6
Network deficiency: 0
Reaction graph of the form
reaction -- reaction label:
s1 -> 2*s1 -- re1
2*s1 -> s1 -- re1r
s1+s2 -> s3 -- re2
s3 -> s1+s2 -- re2r
s3 -> 2*s2 -- re3
2*s2 -> s3 -- re3r
By the Deficiency Zero Theorem, there exists within each positive
stoichiometric compatibility class precisely one equilibrium.
Thus, multiple equilibria cannot exist for the network.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Network satisfies one of the low deficiency theorems.
One should not run the optimization-based methods.
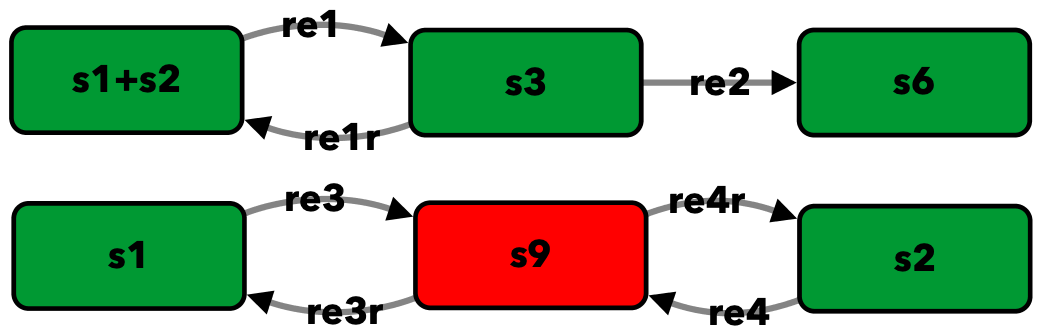
Figure 1Aii of [OMYS17]¶
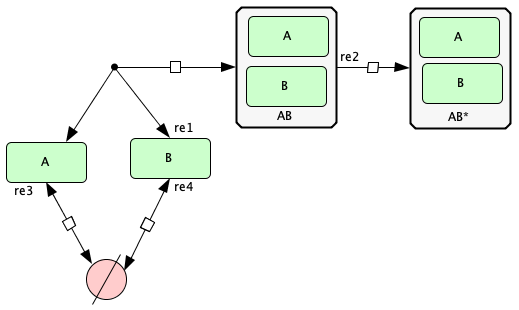
To run this example download the SBML file and script
run_fig1Aii. After running this script we obtain the following output:
Number of species: 4
Number of complexes: 6
Number of reactions: 7
Network deficiency: 0
Reaction graph of the form
reaction -- reaction label:
s1+s2 -> s3 -- re1
s3 -> s1+s2 -- re1r
s3 -> s6 -- re2
s1 -> s9 -- re3
s9 -> s1 -- re3r
s2 -> s9 -- re4
s9 -> s2 -- re4r
By the Deficiency Zero Theorem, the differential equations
cannot admit a positive equilibrium or a cyclic composition
trajectory containing a positive composition. Thus, multiple
equilibria cannot exist for the network.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Network satisfies one of the low deficiency theorems.
One should not run the optimization-based methods.
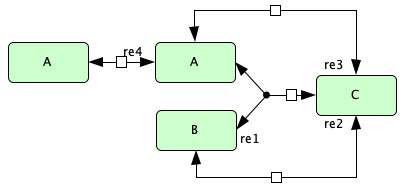
Example 3.D.3 of [Fei79]¶
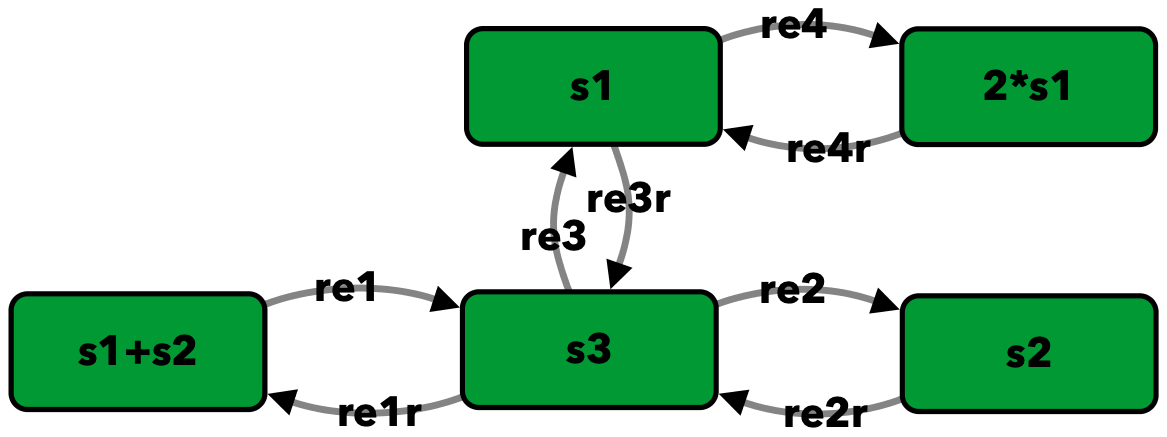
To run this example download the SBML file and script
run_feinberg_ex_3_D_3. After running this script we
obtain the following output:
Number of species: 3
Number of complexes: 5
Number of reactions: 8
Network deficiency: 1
Reaction graph of the form
reaction -- reaction label:
s1+s2 -> s3 -- re1
s3 -> s1+s2 -- re1r
s3 -> s2 -- re2
s2 -> s3 -- re2r
s3 -> s1 -- re3
s1 -> s3 -- re3r
s1 -> 2*s1 -- re4
2*s1 -> s1 -- re4r
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
By the Deficiency One Theorem, the differential equations
admit precisely one equilibrium in each positive stoichiometric
compatibility class. Thus, multiple equilibria cannot exist
for the network.
Network satisfies one of the low deficiency theorems.
One should not run the optimization-based methods.
Mass Conservation Approach¶
Closed graph of Figure 5A of [OMYS17]¶
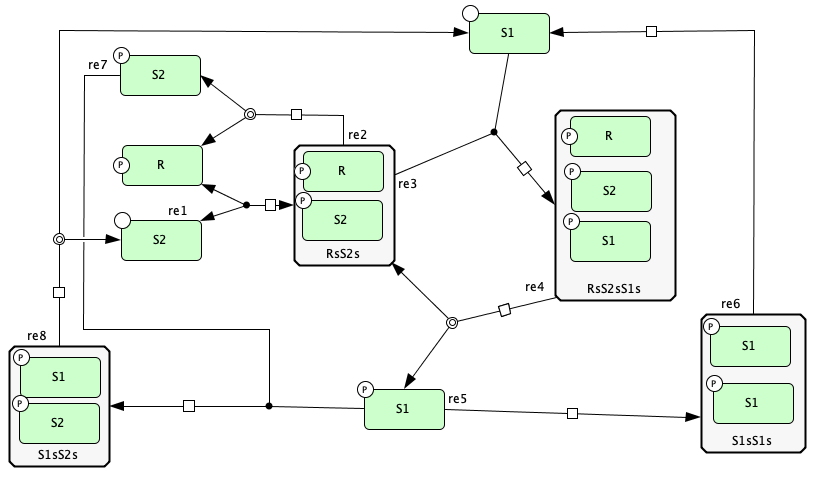
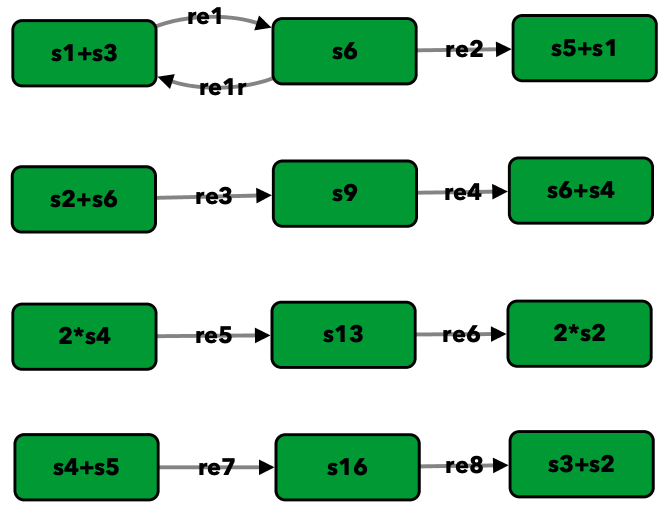
To run this example download the SBML file and script
run_closed_fig5A. After running this script we obtain the
following output:
Number of species: 9
Number of complexes: 12
Number of reactions: 9
Network deficiency: 2
Reaction graph of the form
reaction -- reaction label:
s1+s3 -> s6 -- re1
s6 -> s1+s3 -- re1r
s6 -> s5+s1 -- re2
s2+s6 -> s9 -- re3
s9 -> s6+s4 -- re4
2*s4 -> s13 -- re5
s13 -> 2*s2 -- re6
s4+s5 -> s16 -- re7
s16 -> s3+s2 -- re8
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 3.3645559999999994
Decision Vector:
[re1, re1r, re2, re3, re4, re5, re6, re7, re8, s3, s2, s4]
Species for concentration bounds:
[s1, s6, s5, s9, s13, s16]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 42.63995385169983
Running the multistart optimization method ...
Elapsed time for multistart method: 109.29019284248352
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 16.06424617767334
Smallest value achieved by objective function: 0.0
15 point(s) passed the optimization criteria.
Number of multistability plots found: 2
Elements in params_for_global_min that produce multistability:
[0, 12]
Gene regulatory network with two mutually repressing genes from [OMYS14]¶
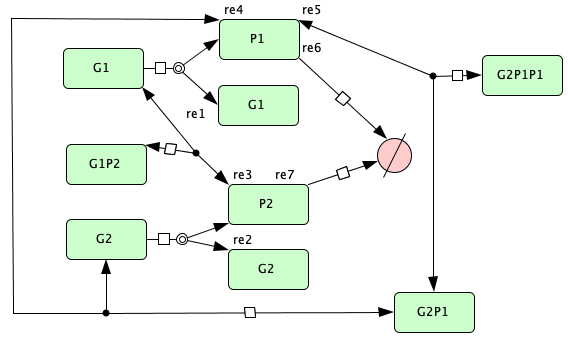
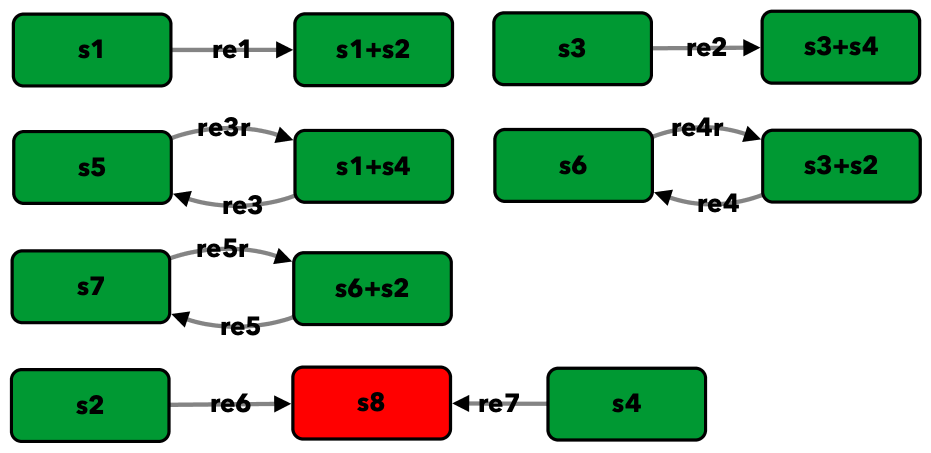
To run this example download the SBML file and script
run_irene2014. After running this script we obtain the following
output:
Number of species: 7
Number of complexes: 13
Number of reactions: 10
Network deficiency: 2
Reaction graph of the form
reaction -- reaction label:
s1 -> s1+s2 -- re1
s3 -> s3+s4 -- re2
s1+s4 -> s5 -- re3
s5 -> s1+s4 -- re3r
s3+s2 -> s6 -- re4
s6 -> s3+s2 -- re4r
s6+s2 -> s7 -- re5
s7 -> s6+s2 -- re5r
s2 -> s8 -- re6
s4 -> s8 -- re7
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 1.772672
Decision Vector:
[re1, re2, re3, re3r, re4, re4r, re5, re5r, re6, re7, s2, s4]
Species for concentration bounds:
[s1, s3, s5, s6, s7]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 25.66311025619507
Running the multistart optimization method ...
Elapsed time for multistart method: 119.89791989326477
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 100.14113593101501
Smallest value achieved by objective function: 0.0
93 point(s) passed the optimization criteria.
Number of multistability plots found: 21
Elements in params_for_global_min that produce multistability:
[1, 3, 9, 11, 15, 21, 24, 27, 32, 35, 40, 45, 56, 62, 70, 79, 80, 83, 84, 85, 88]
Enzymatic reaction with inhibition by substrate from [OMBA09]¶
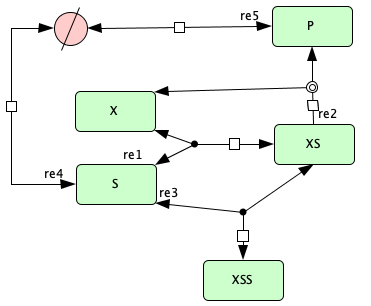
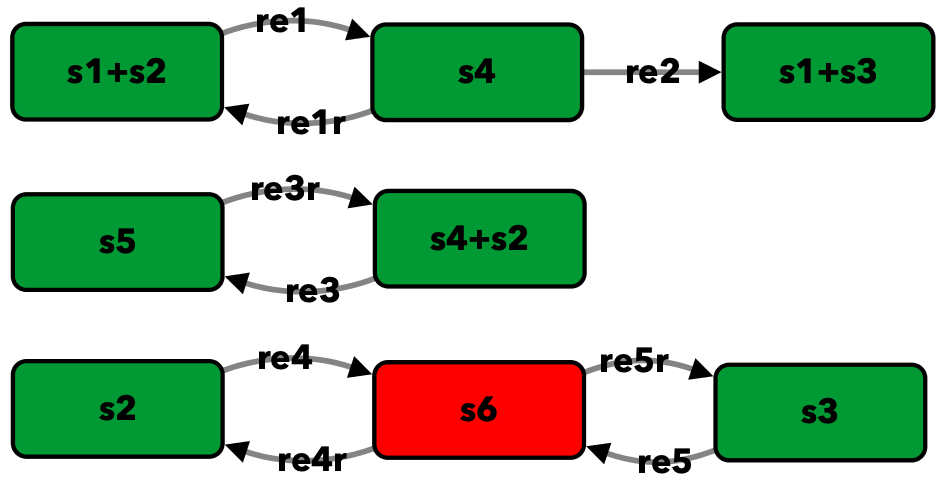
To run this example download the SBML file and script
run_irene2009. After running this script we obtain the following
output:
Number of species: 5
Number of complexes: 8
Number of reactions: 9
Network deficiency: 1
Reaction graph of the form
reaction -- reaction label:
s1+s2 -> s4 -- re1
s4 -> s1+s2 -- re1r
s4 -> s1+s3 -- re2
s4+s2 -> s5 -- re3
s5 -> s4+s2 -- re3r
s2 -> s6 -- re4
s6 -> s2 -- re4r
s3 -> s6 -- re5
s6 -> s3 -- re5r
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 0.715592
Decision Vector:
[re1, re1r, re2, re3, re3r, re4, re4r, re5, re5r, s2]
Species for concentration bounds:
[s1, s4, s3, s5]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 15.607332229614258
Running the multistart optimization method ...
Elapsed time for multistart method: 66.42637610435486
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 72.26282095909119
Smallest value achieved by objective function: 0.0
84 point(s) passed the optimization criteria.
Number of multistability plots found: 48
Elements in params_for_global_min that produce multistability:
[3, 4, 5, 8, 9, 10, 11, 12, 13, 17, 18, 19, 21, 22, 23, 27, 30, 31, 34, 35, 36, 37, 38, 39, 41, 42, 47, 48, 50, 51, 54, 55, 56, 57, 59, 60, 61, 64, 65, 66, 68, 69, 72, 73, 74, 75, 77, 83]
Enzymatic reaction with simple substrate cycle from [HC87]¶
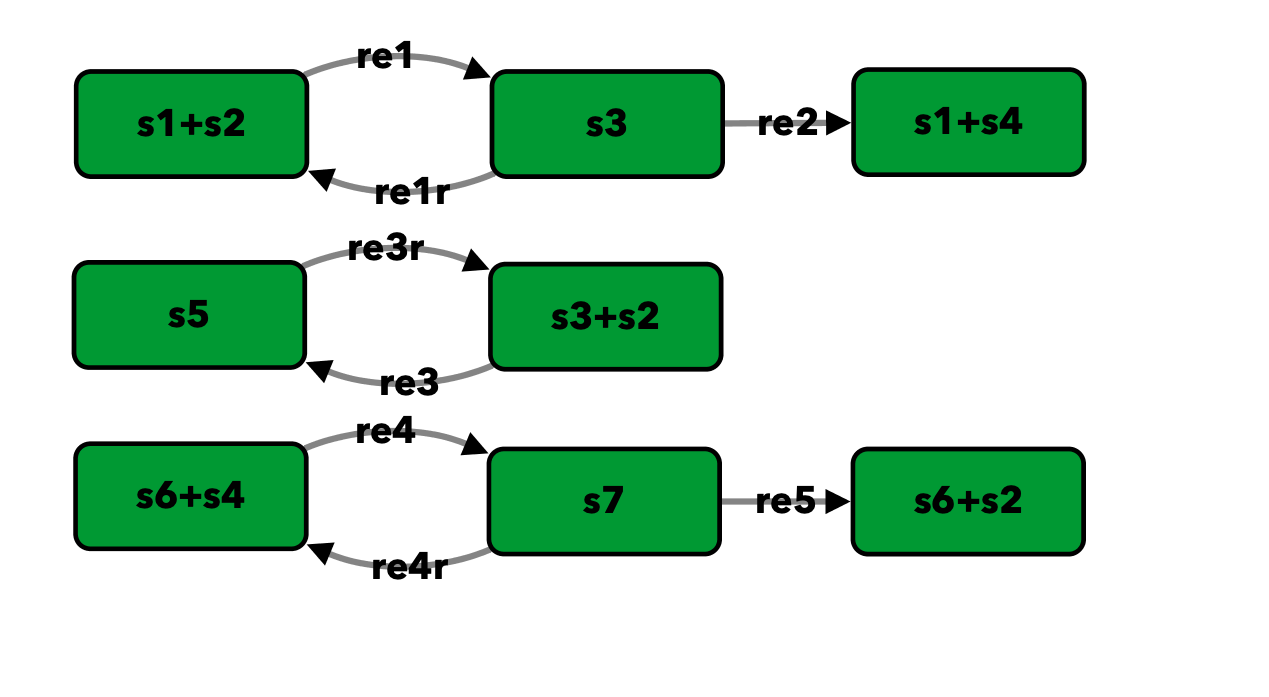
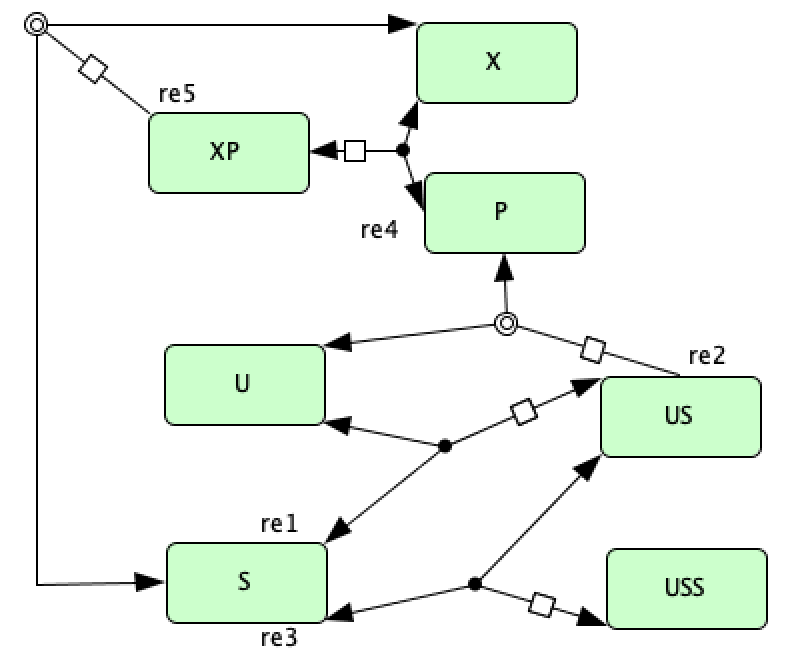
To run this example download the SBML file and script
run_hervagault_canu. After running this script we obtain
the following output:
Number of species: 7
Number of complexes: 8
Number of reactions: 8
Network deficiency: 1
Reaction graph of the form
reaction -- reaction label:
s1+s2 -> s3 -- re1
s3 -> s1+s2 -- re1r
s3 -> s1+s4 -- re2
s3+s2 -> s5 -- re3
s5 -> s3+s2 -- re3r
s6+s4 -> s7 -- re4
s7 -> s6+s4 -- re4r
s7 -> s6+s2 -- re5
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 0.7393859999999997
Decision Vector:
[re1, re1r, re2, re3, re3r, re4, re4r, re5, s2, s6, s7]
Species for concentration bounds:
[s1, s3, s4, s5]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 13.359651803970337
Running the multistart optimization method ...
Elapsed time for multistart method: 103.19853806495667
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 90.50077891349792
Smallest value achieved by objective function: 0.0
96 point(s) passed the optimization criteria.
Number of multistability plots found: 14
Elements in params_for_global_min that produce multistability:
[1, 22, 25, 33, 37, 42, 51, 53, 57, 58, 59, 64, 74, 87]
G1/S transition in the cell cycle of Saccharomyces cerevisiae from [CFRS07]¶
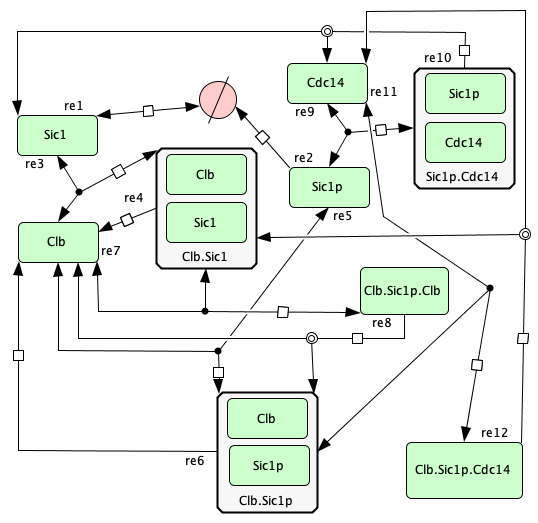
To run this example download the SBML file and script
run_conradi2007. After running this
script we obtain the following output:
Number of species: 9
Number of complexes: 17
Number of reactions: 18
Network deficiency: 5
Reaction graph of the form
reaction -- reaction label:
s1 -> s2 -- re1
s2 -> s1 -- re1r
s3 -> s2 -- re2
s4+s1 -> s5 -- re3
s5 -> s4+s1 -- re3r
s5 -> s4 -- re4
s4+s3 -> s8 -- re5
s8 -> s4+s3 -- re5r
s8 -> s4 -- re6
s5+s4 -> s11 -- re7
s11 -> s5+s4 -- re7r
s11 -> s8+s4 -- re8
s3+s12 -> s13 -- re9
s13 -> s3+s12 -- re9r
s13 -> s1+s12 -- re10
s8+s12 -> s16 -- re11
s16 -> s8+s12 -- re11r
s16 -> s5+s12 -- re12
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 260.415536
Decision Vector:
[re1, re1r, re2, re3, re3r, re4, re5, re5r, re6, re7, re7r, re8, re9, re9r, re10, re11, re11r, re12, s4, s12]
Species for concentration bounds:
[s1, s3, s5, s8, s11, s13, s16]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 73.16450190544128
Running the multistart optimization method ...
Elapsed time for multistart method: 800.0220079421997
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 15.878800868988037
Smallest value achieved by objective function: 0.0
13 point(s) passed the optimization criteria.
Number of multistability plots found: 11
Elements in params_for_global_min that produce multistability:
[0, 1, 2, 3, 5, 6, 7, 8, 10, 11, 12]
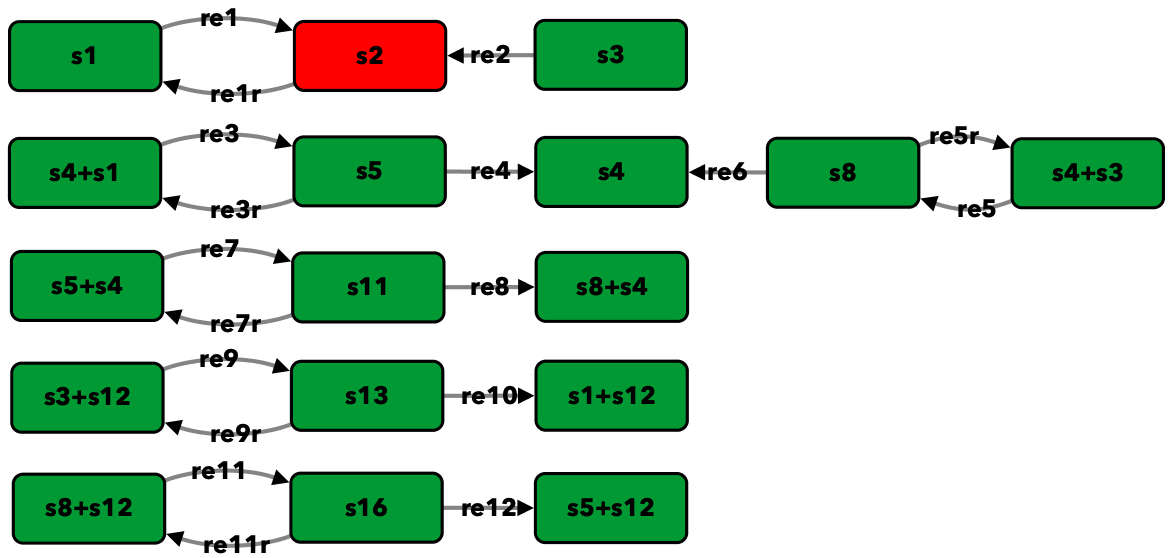
Double phosphorylation in signal transduction of [OGM+06]¶
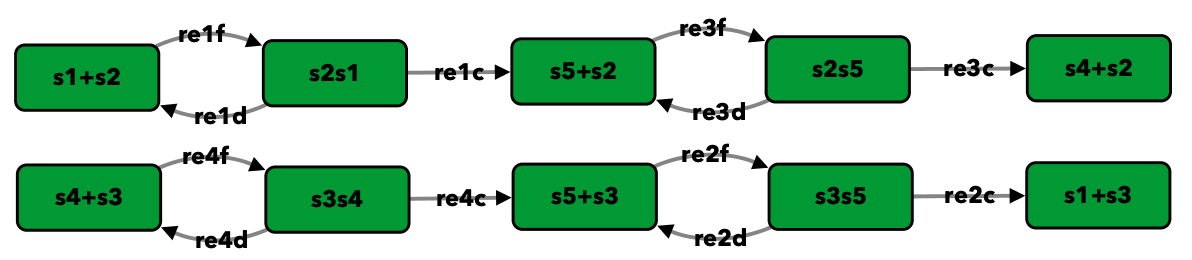
To run this example download the SBML file and script
run_double_phos.
After running this script we obtain the following output:
Number of species: 9
Number of complexes: 10
Number of reactions: 12
Network deficiency: 2
Reaction graph of the form
reaction -- reaction label:
s1+s2 -> s2s1 -- re1f
s2s1 -> s1+s2 -- re1d
s2s1 -> s5+s2 -- re1c
s5+s3 -> s3s5 -- re2f
s3s5 -> s5+s3 -- re2d
s3s5 -> s1+s3 -- re2c
s5+s2 -> s2s5 -- re3f
s2s5 -> s5+s2 -- re3d
s2s5 -> s4+s2 -- re3c
s4+s3 -> s3s4 -- re4f
s3s4 -> s4+s3 -- re4d
s3s4 -> s5+s3 -- re4c
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 5.184272
Decision Vector:
[re1f, re1d, re1c, re2f, re2d, re2c, re3f, re3d, re3c, re4f, re4d, re4c, s2, s3, s3s4]
Species for concentration bounds:
[s1, s5, s2s1, s3s5, s4, s2s5]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 18.401470184326172
Running the multistart optimization method ...
Elapsed time for multistart method: 95.46931576728821
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 372.1889531612396
Smallest value achieved by objective function: 0.0
97 point(s) passed the optimization criteria.
Number of multistability plots found: 89
Elements in params_for_global_min that produce multistability:
[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60,
61, 62, 64, 65, 66, 67, 69, 70, 71, 72, 73, 74, 75, 76, 77, 79, 80, 81, 82, 83, 84, 87, 88, 90, 91, 92, 93, 94, 95, 96]
Double insulin binding¶
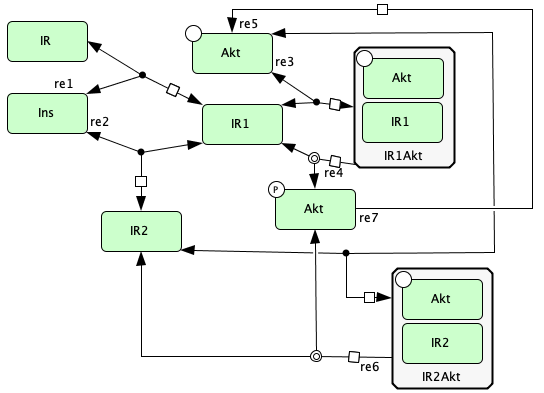
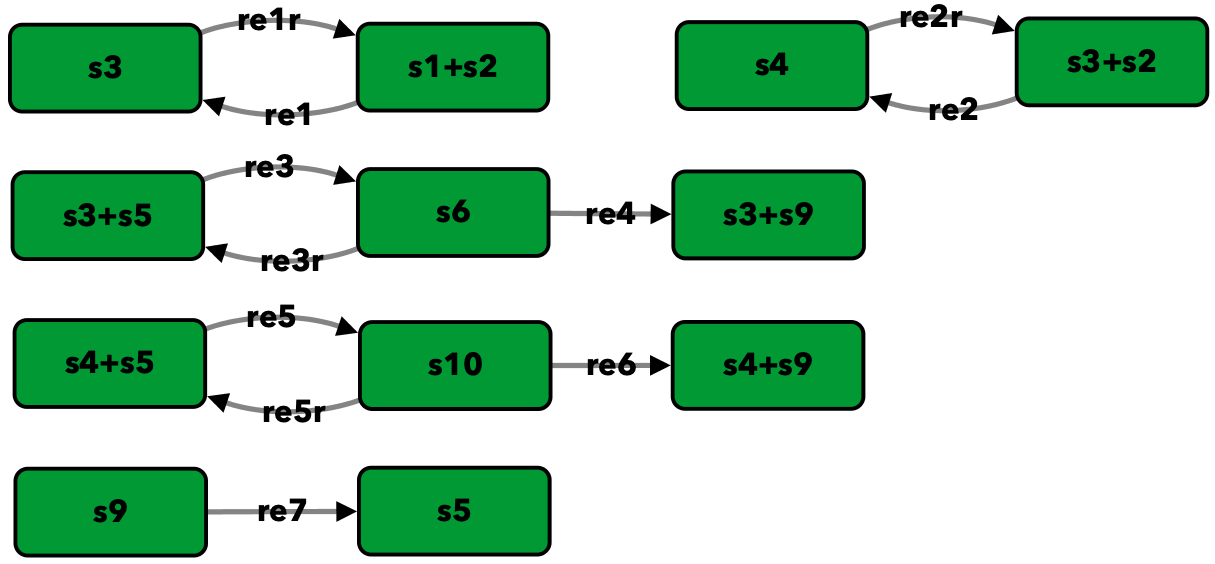
To run this example download the SBML file and script
run_double_insulin_binding.
After running this script we obtain the following output:
Number of species: 8
Number of complexes: 12
Number of reactions: 11
Network deficiency: 2
Reaction graph of the form
reaction -- reaction label:
s1+s2 -> s3 -- re1
s3 -> s1+s2 -- re1r
s3+s2 -> s4 -- re2
s4 -> s3+s2 -- re2r
s3+s5 -> s6 -- re3
s6 -> s3+s5 -- re3r
s6 -> s3+s9 -- re4
s4+s5 -> s10 -- re5
s10 -> s4+s5 -- re5r
s10 -> s4+s9 -- re6
s9 -> s5 -- re7
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 2.2847300000000006
Decision Vector:
[re1, re1r, re2, re2r, re3, re3r, re4, re5, re5r, re6, re7, s2, s5, s10]
Species for concentration bounds:
[s1, s3, s4, s6, s9]
Running feasible point method for 100 iterations ...
Elapsed time for feasible point method: 25.920205116271973
Running the multistart optimization method ...
Elapsed time for multistart method: 94.97992706298828
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 652.6215398311615
Smallest value achieved by objective function: 2.3317319454459066e-31
67 point(s) passed the optimization criteria.
Number of multistability plots found: 2
Elements in params_for_global_min that produce multistability:
[8, 38]
p85-p110-PTEN¶
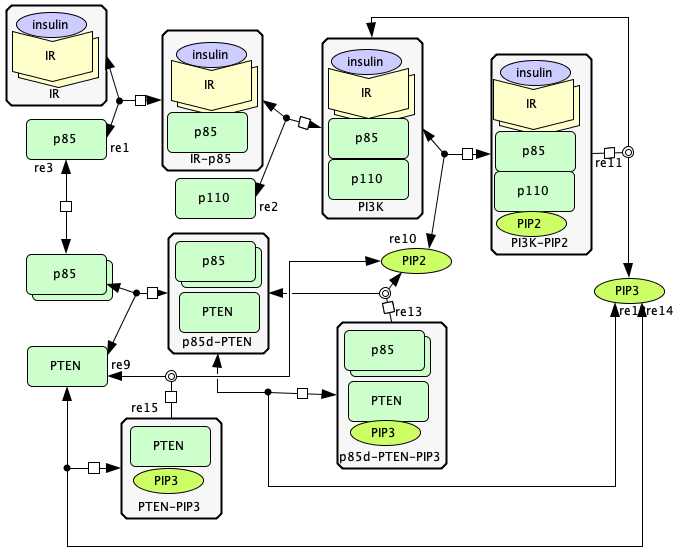
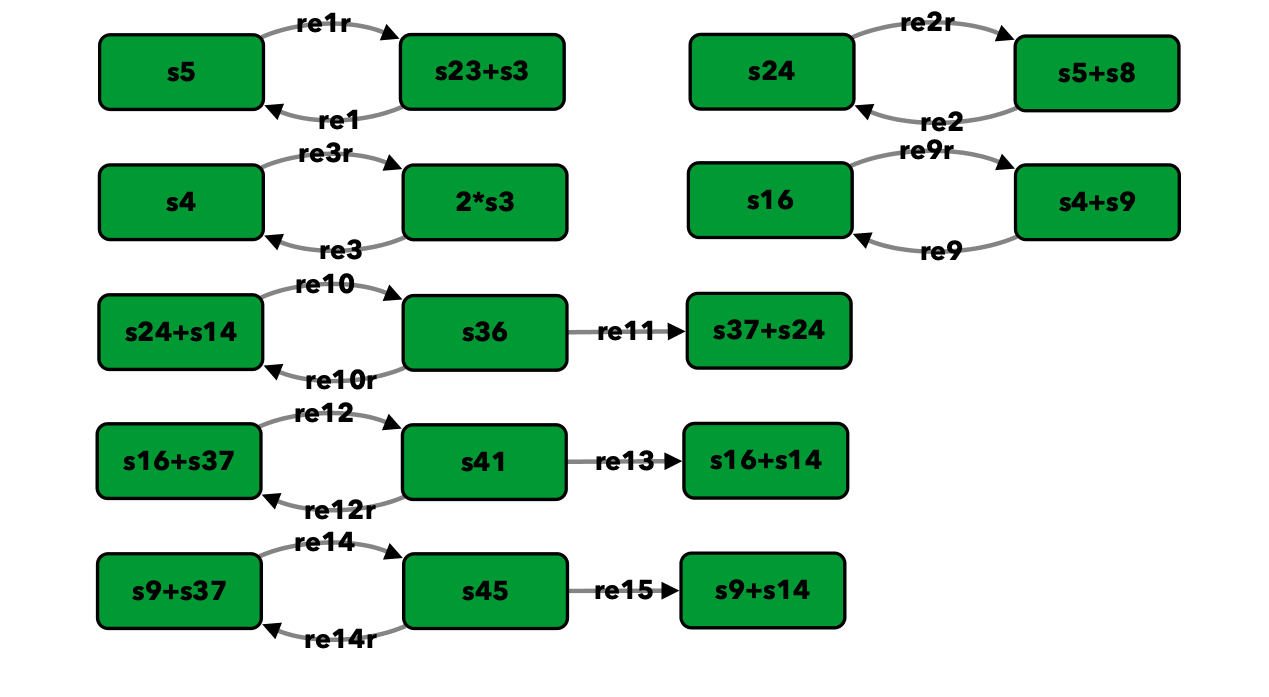
To run this example download the SBML file and script
run_p85-p110-PTEN. After running this script using four cores,
we obtain the following output (for more information on running this script in parallel see Parallel CRNT4SBML):
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 107.71943200000001
Elapsed time for creating Equilibrium Manifold: 108.786772
Elapsed time for creating Equilibrium Manifold: 108.861678
Elapsed time for creating Equilibrium Manifold: 109.171994
Running feasible point method for 5000 iterations ...
Elapsed time for feasible point method: 2519.281478
Running the multistart optimization method ...
Elapsed time for multistart method: 403.41574900000023
Number of species: 13
Number of complexes: 17
Number of reactions: 17
Network deficiency: 2
Reaction graph of the form
reaction -- reaction label:
s23+s3 -> s5 -- re1
s5 -> s23+s3 -- re1r
s5+s8 -> s24 -- re2
s24 -> s5+s8 -- re2r
2*s3 -> s4 -- re3
s4 -> 2*s3 -- re3r
s4+s9 -> s16 -- re9
s16 -> s4+s9 -- re9r
s24+s14 -> s36 -- re10
s36 -> s24+s14 -- re10r
s36 -> s37+s24 -- re11
s16+s37 -> s41 -- re12
s41 -> s16+s37 -- re12r
s41 -> s16+s14 -- re13
s9+s37 -> s45 -- re14
s45 -> s9+s37 -- re14r
s45 -> s9+s14 -- re15
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision Vector:
[re1, re1r, re2, re2r, re3, re3r, re9, re9r, re10, re10r, re11, re12, re12r, re13, re14, re14r, re15, s3, s8, s9, s14, s37]
Species for concentration bounds:
[s23, s5, s24, s4, s16, s36, s41, s45]
A parallel version of numerical continuation is not available.
Numerical continuation will be ran using only one core.
For your convenience, the provided parameters have been saved in the current directory under the name params.npy.
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 5766.086745023727
Smallest value achieved by objective function: 0.0
429 point(s) passed the optimization criteria.
Number of multistability plots found: 5
Elements in params_for_global_min that produce multistability:
[171, 191, 213, 272, 296]
Closed version of Figure 4B from [OMYS17]¶
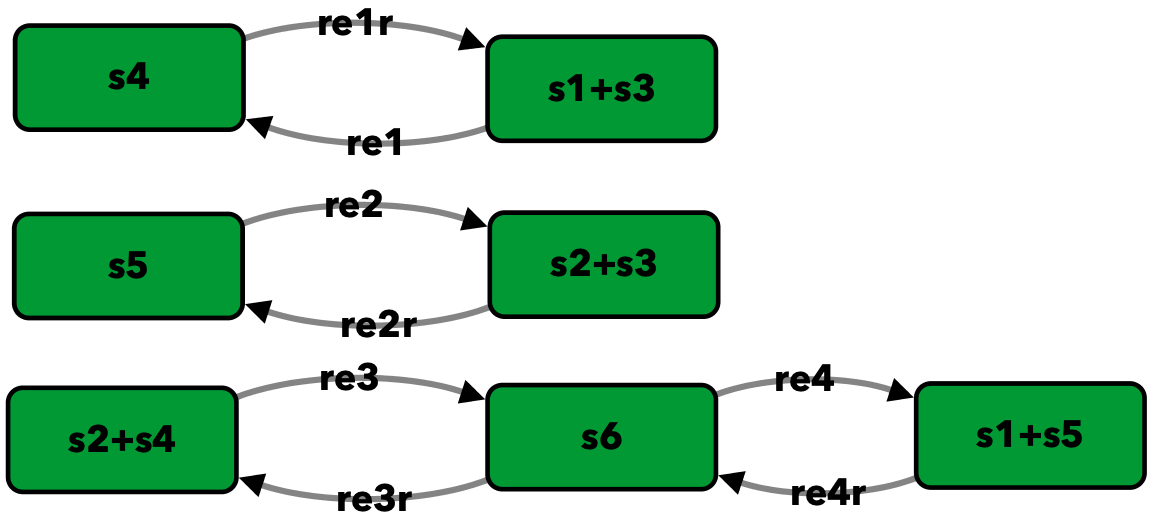
To run this example download the SBML file and script
run_Fig4B_closed. After running this script using four cores,
we obtain the following output (for more information on running this script in parallel see Parallel CRNT4SBML):
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 1.2114520000000004
Elapsed time for creating Equilibrium Manifold: 1.2372060000000005
Elapsed time for creating Equilibrium Manifold: 1.229298
Elapsed time for creating Equilibrium Manifold: 1.2412400000000003
Running feasible point method for 10000 iterations ...
Elapsed time for feasible point method: 518.759626
Running the multistart optimization method ...
Elapsed time for multistart method: 2561.635341
Number of species: 6
Number of complexes: 7
Number of reactions: 8
Network deficiency: 1
Reaction graph of the form
reaction -- reaction label:
s1+s3 -> s4 -- re1
s4 -> s1+s3 -- re1r
s5 -> s2+s3 -- re2
s2+s3 -> s5 -- re2r
s2+s4 -> s6 -- re3
s6 -> s2+s4 -- re3r
s6 -> s1+s5 -- re4
s1+s5 -> s6 -- re4r
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision Vector:
[re1, re1r, re2, re2r, re3, re3r, re4, re4r, s3, s5, s2]
Species for concentration bounds:
[s1, s4, s6]
Smallest value achieved by objective function: 2.454796889817468e-10
0 point(s) passed the optimization criteria.
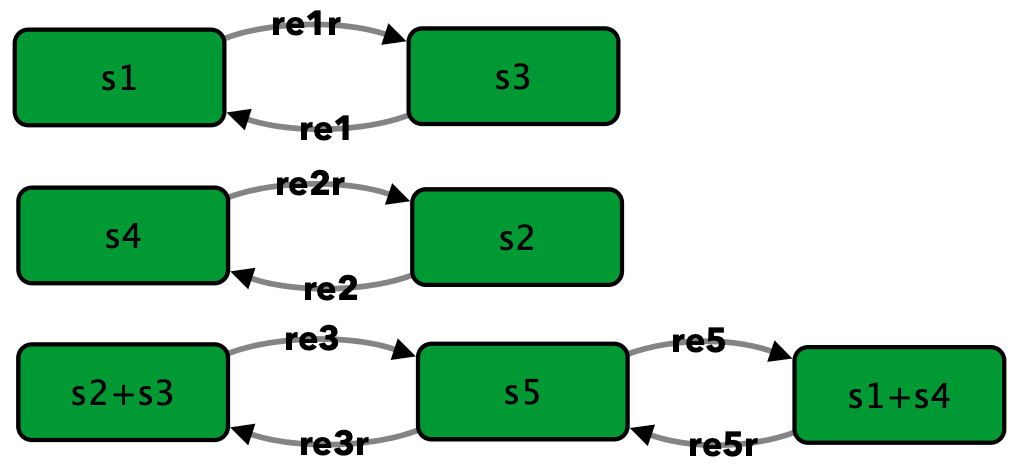
Closed version of Figure 4C from [OMYS17]¶
To run this example download the SBML file and script
run_Fig4C_closed. After running this script using four cores,
we obtain the following output (for more information on running this script in parallel see Parallel CRNT4SBML):
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Creating Equilibrium Manifold ...
Elapsed time for creating Equilibrium Manifold: 0.9796280000000004
Elapsed time for creating Equilibrium Manifold: 0.9905299999999997
Elapsed time for creating Equilibrium Manifold: 0.997398
Elapsed time for creating Equilibrium Manifold: 0.9981960000000001
Running feasible point method for 10000 iterations ...
Elapsed time for feasible point method: 236.957728
Running the multistart optimization method ...
Elapsed time for multistart method: 2115.088291
Number of species: 5
Number of complexes: 7
Number of reactions: 8
Network deficiency: 1
Reaction graph of the form
reaction -- reaction label:
s3 -> s1 -- re1
s1 -> s3 -- re1r
s2 -> s4 -- re2
s4 -> s2 -- re2r
s2+s3 -> s5 -- re3
s5 -> s2+s3 -- re3r
s5 -> s1+s4 -- re5
s1+s4 -> s5 -- re5r
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision Vector:
[re1, re1r, re2, re2r, re3, re3r, re5, re5r, s2, s4]
Species for concentration bounds:
[s3, s1, s5]
Smallest value achieved by objective function: 1.2913762450176939e-09
0 point(s) passed the optimization criteria.
Semi-diffusive Approach¶
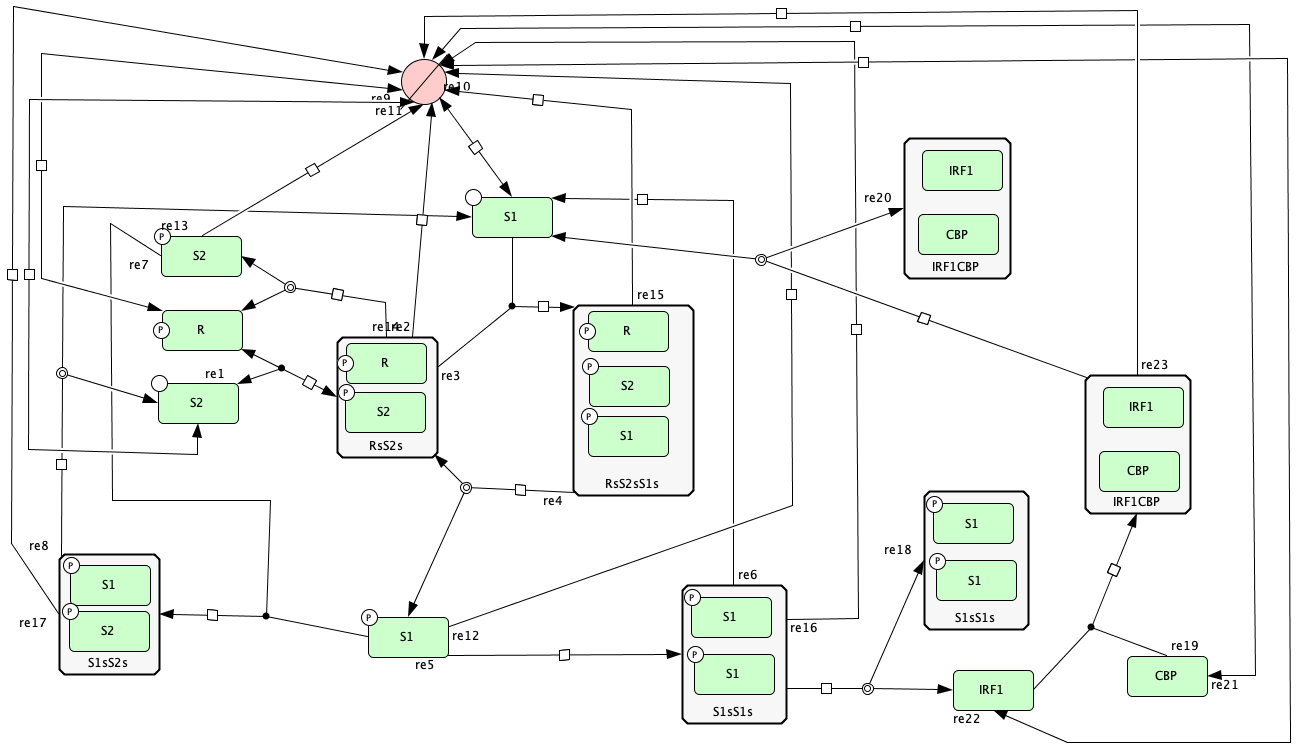
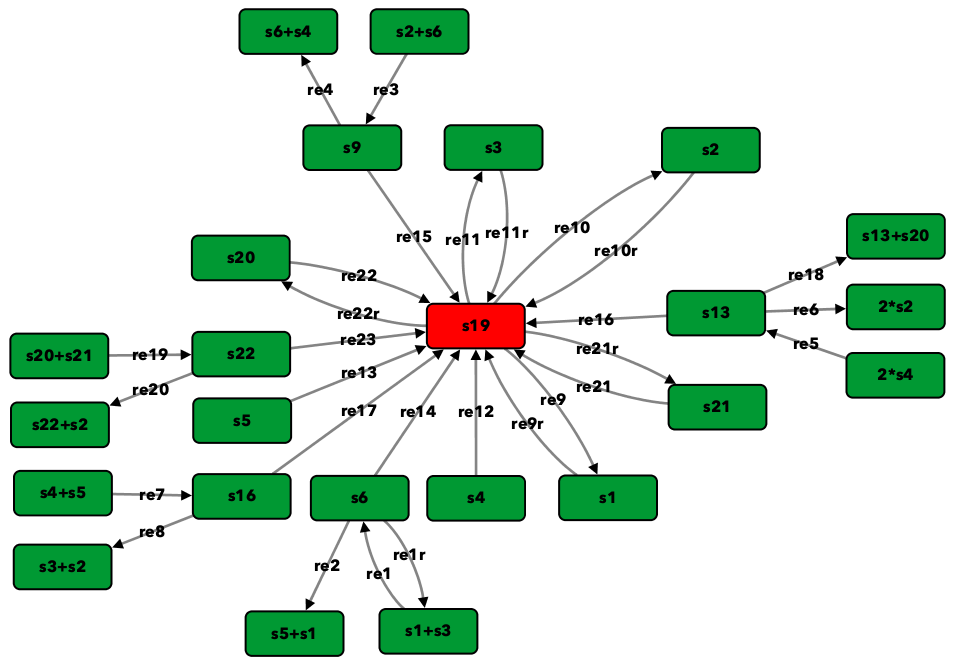
Figure 5B of [OMYS17]¶
To run this example download the SBML file and script
run_open_fig5B. After running this script we obtain the
following output:
Number of species: 12
Number of complexes: 24
Number of reactions: 29
Network deficiency: 11
Reaction graph of the form
reaction -- reaction label:
s1+s3 -> s6 -- re1
s6 -> s1+s3 -- re1r
s6 -> s5+s1 -- re2
s2+s6 -> s9 -- re3
s9 -> s6+s4 -- re4
2*s4 -> s25 -- re5
s25 -> 2*s2 -- re6
s4+s5 -> s16 -- re7
s16 -> s3+s2 -- re8
s19 -> s1 -- re9
s1 -> s19 -- re9r
s19 -> s2 -- re10
s2 -> s19 -- re10r
s19 -> s3 -- re11
s3 -> s19 -- re11r
s4 -> s19 -- re12
s5 -> s19 -- re13
s6 -> s19 -- re14
s9 -> s19 -- re15
s25 -> s19 -- re16
s16 -> s19 -- re17
s25 -> s25+s20 -- re18
s20+s21 -> s22 -- re19
s22 -> s22+s2 -- re20
s21 -> s19 -- re21
s19 -> s21 -- re21r
s20 -> s19 -- re22
s19 -> s20 -- re22r
s22 -> s19 -- re23
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision vector for optimization:
[v_2, v_3, v_4, v_5, v_6, v_8, v_11, v_13, v_15, v_18, v_20, v_21, v_22, v_24, v_25, v_27, v_29]
Reaction labels for decision vector:
['re1r', 're2', 're3', 're4', 're5', 're7', 're9r', 're10r', 're11r', 're14', 're16', 're17', 're18', 're20', 're21', 're22', 're23']
Key species:
['s1', 's3', 's2', 's20', 's21']
Non key species:
['s6', 's5', 's9', 's4', 's25', 's16', 's22']
Boundary species:
['s19']
Running feasible point method for 50 iterations ...
Elapsed time for feasible point method: 14.352675676345825
Running the multistart optimization method ...
Elapsed time for multistart method: 352.3979892730713
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 42.74703788757324
Smallest value achieved by objective function: 0.0
22 point(s) passed the optimization criteria.
Number of multistability plots found: 4
Elements in params_for_global_min that produce multistability:
[0, 1, 3, 16]
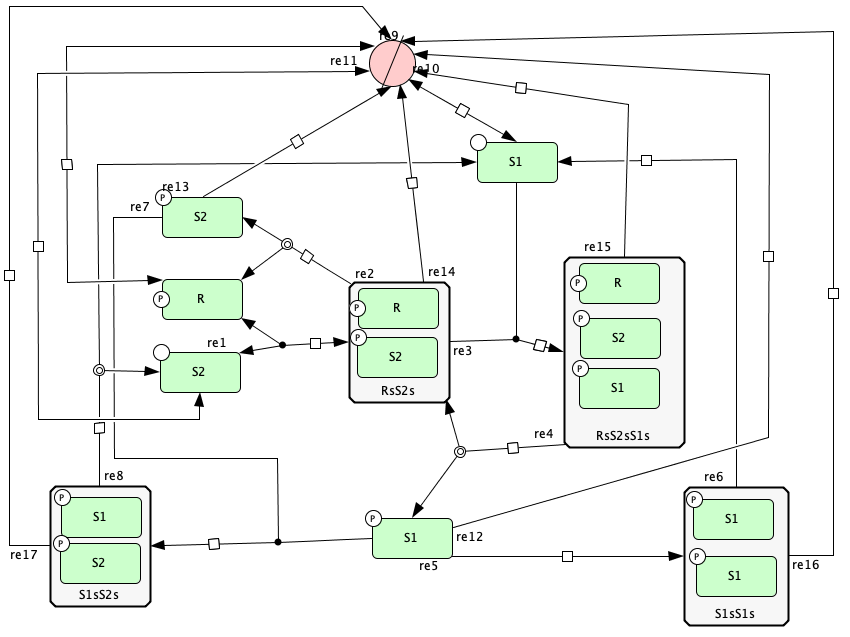
Open version of Figure 5A from [OMYS17]¶
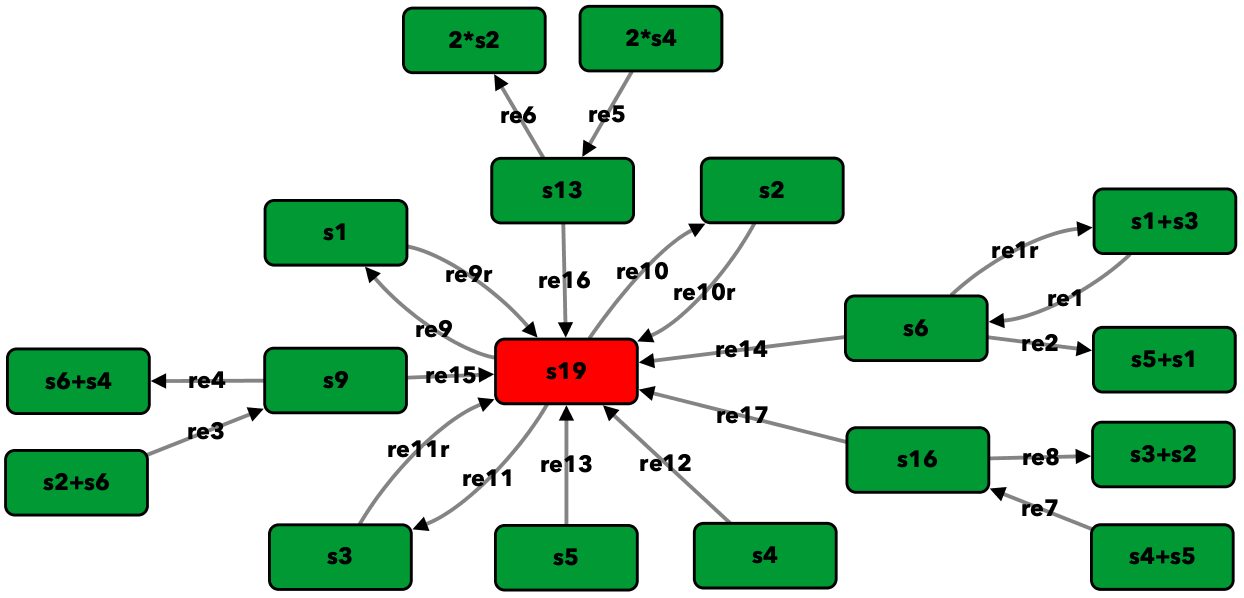
To run this example download the SBML file and script
run_open_fig5A. After running this script we obtain the
following output:
Number of species: 9
Number of complexes: 18
Number of reactions: 21
Network deficiency: 8
Reaction graph of the form
reaction -- reaction label:
s1+s3 -> s6 -- re1
s6 -> s1+s3 -- re1r
s6 -> s5+s1 -- re2
s2+s6 -> s9 -- re3
s9 -> s6+s4 -- re4
2*s4 -> s13 -- re5
s13 -> 2*s2 -- re6
s4+s5 -> s16 -- re7
s16 -> s3+s2 -- re8
s19 -> s1 -- re9
s1 -> s19 -- re9r
s19 -> s2 -- re10
s2 -> s19 -- re10r
s19 -> s3 -- re11
s3 -> s19 -- re11r
s4 -> s19 -- re12
s5 -> s19 -- re13
s6 -> s19 -- re14
s9 -> s19 -- re15
s13 -> s19 -- re16
s16 -> s19 -- re17
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision vector for optimization:
[v_2, v_3, v_4, v_5, v_6, v_8, v_11, v_13, v_15, v_18, v_20, v_21]
Reaction labels for decision vector:
['re1r', 're2', 're3', 're4', 're5', 're7', 're9r', 're10r', 're11r', 're14', 're16', 're17']
Key species:
['s1', 's3', 's2']
Non key species:
['s6', 's5', 's9', 's4', 's13', 's16']
Boundary species:
['s19']
Running feasible point method for 500 iterations ...
Elapsed time for feasible point method: 40.84808683395386
Running the multistart optimization method ...
Elapsed time for multistart method: 597.4433598518372
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 1777.679843902588
Smallest value achieved by objective function: 0.0
108 point(s) passed the optimization criteria.
Number of multistability plots found: 1
Elements in params_for_global_min that produce multistability:
[85]
Figure 4B from [OMYS17]¶
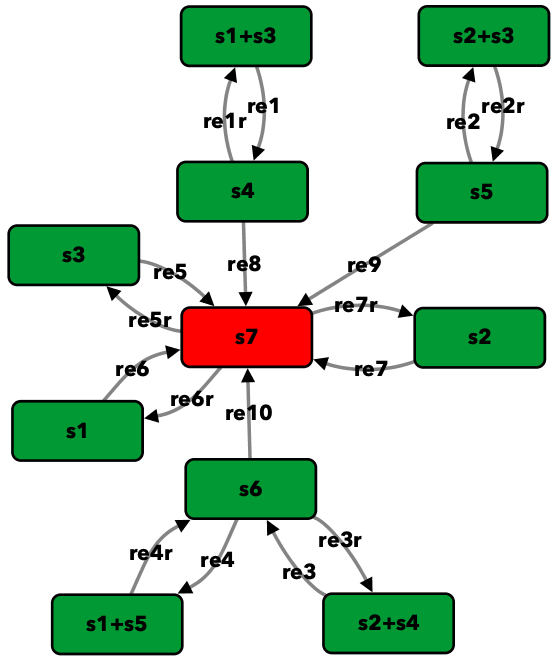
To run this example download the SBML file and script
run_Fig4B_open. After running this script using four cores,
we obtain the following output (for more information on running this script in parallel see Parallel CRNT4SBML):
Running feasible point method for 10000 iterations ...
Elapsed time for feasible point method: 73.587205
Running the multistart optimization method ...
Elapsed time for multistart method: 3675.938109
Number of species: 6
Number of complexes: 11
Number of reactions: 17
Network deficiency: 4
Reaction graph of the form
reaction -- reaction label:
s1+s3 -> s4 -- re1
s4 -> s1+s3 -- re1r
s5 -> s2+s3 -- re2
s2+s3 -> s5 -- re2r
s2+s4 -> s6 -- re3
s6 -> s2+s4 -- re3r
s6 -> s1+s5 -- re4
s1+s5 -> s6 -- re4r
s3 -> s7 -- re5
s7 -> s3 -- re5r
s1 -> s7 -- re6
s7 -> s1 -- re6r
s2 -> s7 -- re7
s7 -> s2 -- re7r
s4 -> s7 -- re8
s5 -> s7 -- re9
s6 -> s7 -- re10
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision vector for optimization:
[v_2, v_4, v_5, v_6, v_7, v_8, v_9, v_11, v_13, v_15, v_16]
Reaction labels for decision vector:
['re1r', 're2r', 're3', 're3r', 're4', 're4r', 're5', 're6', 're7', 're8', 're9']
Key species:
['s1', 's3', 's2']
Non key species:
['s4', 's5', 's6']
Boundary species:
['s7']
Smallest value achieved by objective function: 2.3045037796933692e-10
0 point(s) passed the optimization criteria.
Figure 4C from [OMYS17]¶
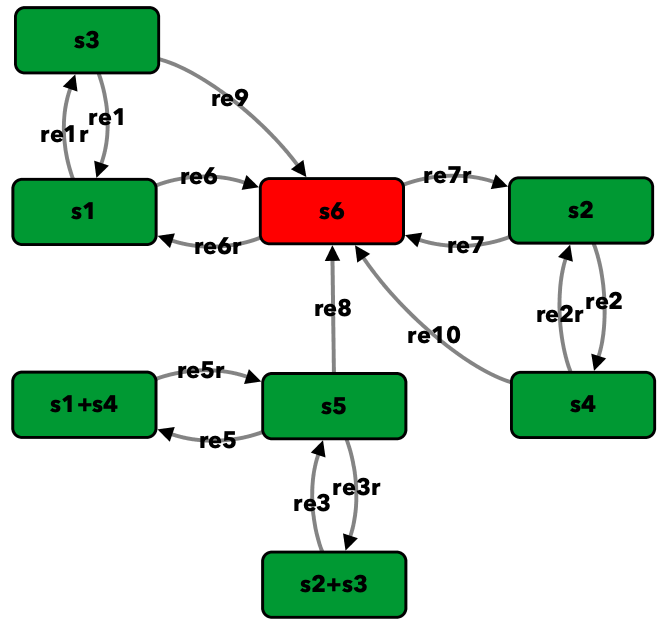
To run this example download the SBML file and script
run_Fig4C_open. After running this script using four cores,
we obtain the following output (for more information on running this script in parallel see Parallel CRNT4SBML):
Running feasible point method for 10000 iterations ...
Elapsed time for feasible point method: 57.548688
Running the multistart optimization method ...
Elapsed time for multistart method: 1432.020307
Number of species: 5
Number of complexes: 8
Number of reactions: 15
Network deficiency: 2
Reaction graph of the form
reaction -- reaction label:
s3 -> s1 -- re1
s1 -> s3 -- re1r
s2 -> s4 -- re2
s4 -> s2 -- re2r
s2+s3 -> s5 -- re3
s5 -> s2+s3 -- re3r
s5 -> s1+s4 -- re5
s1+s4 -> s5 -- re5r
s1 -> s6 -- re6
s6 -> s1 -- re6r
s2 -> s6 -- re7
s6 -> s2 -- re7r
s5 -> s6 -- re8
s3 -> s6 -- re9
s4 -> s6 -- re10
The network does not satisfy the Deficiency Zero Theorem, multistability cannot be excluded.
The network does not satisfy the Deficiency One Theorem, multistability cannot be excluded.
Decision vector for optimization:
[v_2, v_4, v_5, v_6, v_7, v_8, v_9, v_11, v_14, v_15]
Reaction labels for decision vector:
['re1r', 're2r', 're3', 're3r', 're5', 're5r', 're6', 're7', 're9', 're10']
Key species:
['s1', 's2']
Non key species:
['s3', 's4', 's5']
Boundary species:
['s6']
Smallest value achieved by objective function: 4.5692676949897973e-10
0 point(s) passed the optimization criteria.
General Approach¶
Song model of [FSW+16]¶
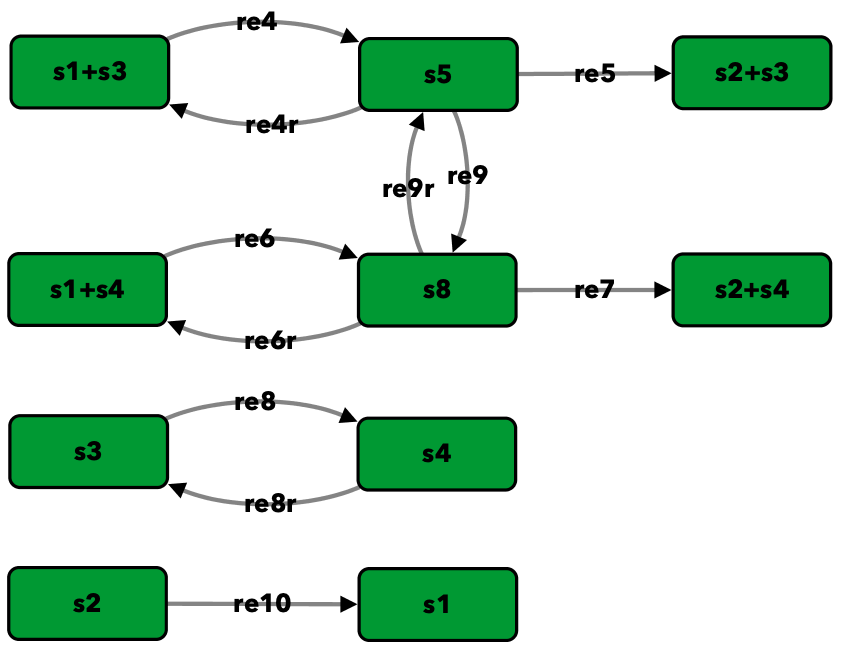
To run this example download the SBML file and script
run_song_model. After running this script we obtain the
following output:
Number of species: 6
Number of complexes: 10
Number of reactions: 11
Network deficiency: 3
Reaction graph of the form
reaction -- reaction label:
s1+s3 -> s5 -- re4
s5 -> s1+s3 -- re4r
s5 -> s2+s3 -- re5
s1+s4 -> s8 -- re6
s8 -> s1+s4 -- re6r
s8 -> s2+s4 -- re7
s3 -> s4 -- re8
s4 -> s3 -- re8r
s5 -> s8 -- re9
s8 -> s5 -- re9r
s2 -> s1 -- re10
[re4, re4r, re5, re6, re6r, re7, re8, re8r, re9, re9r, re10, s1, s3, s5, s2, s4, s8]
Running the multistart optimization method ...
Elapsed time for multistart method: 1228.3208582401276
Running continuity analysis ...
Elapsed time for continuity analysis in seconds: 28.140807151794434
Smallest value achieved by objective function: 0.0
5 point(s) passed the optimization criteria.
Number of multistability plots found: 2
Elements in params_for_global_min that produce multistability:
[1, 4]